When a progress curve is linear from the beginning, and remains linear throughout the experiment, a single end point reading can approximate the reaction rate. However, frequently when interrogating protein kinases and their inhibitors, there is a delay in the onset of the reaction, or other changes in reaction rate over time. In these cases, important information about the reaction can be missed or mischaracterized in an endpoint assay. Using a continuous assay format, like our PhosphoSens® technology, will visualize and quantify enzyme activity with confidence.
While visual inspection is the most common method of determining the range of the progress curve from which to extract rate information, this method is not practical when thousands of progress curves are generated in an experimental day. An automated method is essential for high throughput assessment of enzyme activity, for example in large-scale lead optimization or selectivity assessment, both of which are essential for developing cutting edge drug candidates.
Our fully automated linear range finding algorithm substantially increases the speed, reliability, and reproducibility of linear range definition for enzyme kinetic studies. The method enables high throughput screening in a kinetic enzyme assay, for example making possible rapid analysis of very large data sets from kinome profiling data.
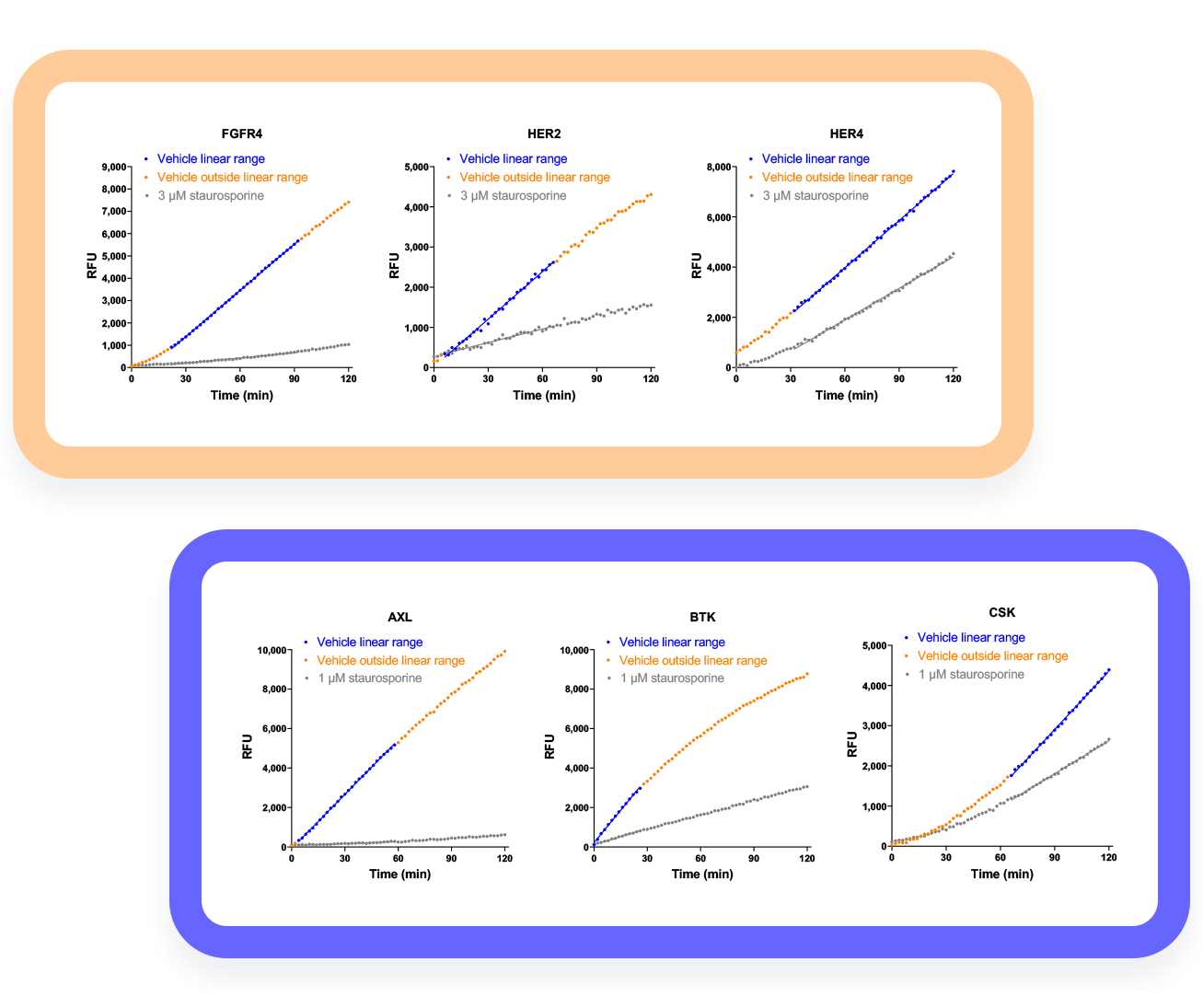
Examples of automated linear range finding applied to kinome profiling.
"Automated Linear Range Determination and Applications to Enzymatic Progress Curves"
Download our scientific poster to learn more about what your data can tell you.
We present:
- A novel automated protocol to streamline and optimize linear range finding.
- A method to characterize inhibition of the enzyme target using a variety of biochemical assessments: simple dose-response, mode of inhibition, time-dependent inhibition, reversibility, residence time for reversible inhibitors, kinact/KI for irreversible inhibitors, and full kinome selectivity profiling.
- Applications for the analysis of protein kinases as a group of high-profile drug targets with 518 distinct members, using the PhosphoSens® assay with data analysis using Scigilian Analyze.